Machine Learning Tutorial: From Data Cleaning to Classification#
In this MACHINE LEARNING tutorial, we will cover a full machine learning pipeline using the breast cancer dataset from scikit-learn for classification task. Here we are creating a model that classifies the features from breast cancer dataset into two classes, 0 or 1 (class labels). To know the basics of Machine Learning, refer here.
# install the ML library, scikit-learn
%pip install scikit-learn
Step 1: Import libraries#
# data anaysis packages
import pandas as pd
import numpy as np
# Plotting packages
import matplotlib.pyplot as plt
import seaborn as sns
#machine learning packages
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.impute import SimpleImputer
from sklearn.ensemble import RandomForestClassifier
from sklearn.metrics import classification_report, confusion_matrix
#Built-in dataset in sci-kit learn (sklearn), breast cancer dataset
from sklearn.datasets import load_breast_cancer
Step 2: Load and explore the data#
# load the dataset and store it in the pandas dataframe df with column names (X) =feature names and target (y) ic column "target"
data = load_breast_cancer()
df = pd.DataFrame(data.data, columns=data.feature_names)
df['target'] = data.target
# Preview the dataset
print(df.head()) # print first 5 rows of the data
mean radius mean texture mean perimeter mean area mean smoothness \
0 17.99 10.38 122.80 1001.0 0.11840
1 20.57 17.77 132.90 1326.0 0.08474
2 19.69 21.25 130.00 1203.0 0.10960
3 11.42 20.38 77.58 386.1 0.14250
4 20.29 14.34 135.10 1297.0 0.10030
mean compactness mean concavity mean concave points mean symmetry \
0 0.27760 0.3001 0.14710 0.2419
1 0.07864 0.0869 0.07017 0.1812
2 0.15990 0.1974 0.12790 0.2069
3 0.28390 0.2414 0.10520 0.2597
4 0.13280 0.1980 0.10430 0.1809
mean fractal dimension ... worst texture worst perimeter worst area \
0 0.07871 ... 17.33 184.60 2019.0
1 0.05667 ... 23.41 158.80 1956.0
2 0.05999 ... 25.53 152.50 1709.0
3 0.09744 ... 26.50 98.87 567.7
4 0.05883 ... 16.67 152.20 1575.0
worst smoothness worst compactness worst concavity worst concave points \
0 0.1622 0.6656 0.7119 0.2654
1 0.1238 0.1866 0.2416 0.1860
2 0.1444 0.4245 0.4504 0.2430
3 0.2098 0.8663 0.6869 0.2575
4 0.1374 0.2050 0.4000 0.1625
worst symmetry worst fractal dimension target
0 0.4601 0.11890 0
1 0.2750 0.08902 0
2 0.3613 0.08758 0
3 0.6638 0.17300 0
4 0.2364 0.07678 0
[5 rows x 31 columns]
find the number of rows and columns, column names, and datatype of columns#
print(df.info())
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 569 entries, 0 to 568
Data columns (total 31 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 mean radius 569 non-null float64
1 mean texture 569 non-null float64
2 mean perimeter 569 non-null float64
3 mean area 569 non-null float64
4 mean smoothness 569 non-null float64
5 mean compactness 569 non-null float64
6 mean concavity 569 non-null float64
7 mean concave points 569 non-null float64
8 mean symmetry 569 non-null float64
9 mean fractal dimension 569 non-null float64
10 radius error 569 non-null float64
11 texture error 569 non-null float64
12 perimeter error 569 non-null float64
13 area error 569 non-null float64
14 smoothness error 569 non-null float64
15 compactness error 569 non-null float64
16 concavity error 569 non-null float64
17 concave points error 569 non-null float64
18 symmetry error 569 non-null float64
19 fractal dimension error 569 non-null float64
20 worst radius 569 non-null float64
21 worst texture 569 non-null float64
22 worst perimeter 569 non-null float64
23 worst area 569 non-null float64
24 worst smoothness 569 non-null float64
25 worst compactness 569 non-null float64
26 worst concavity 569 non-null float64
27 worst concave points 569 non-null float64
28 worst symmetry 569 non-null float64
29 worst fractal dimension 569 non-null float64
30 target 569 non-null int32
dtypes: float64(30), int32(1)
memory usage: 135.7 KB
None
The data has 569 rows and 31 columns
except “target” column, all the columns are float
Step 3: Data cleaning#
# to check for missing values in each column
print(df.isnull().sum())
# Use imputer if missing values were present
# imputer = SimpleImputer(strategy="mean")
# df.iloc[:, :-1] = imputer.fit_transform(df.iloc[:, :-1])
mean radius 0
mean texture 0
mean perimeter 0
mean area 0
mean smoothness 0
mean compactness 0
mean concavity 0
mean concave points 0
mean symmetry 0
mean fractal dimension 0
radius error 0
texture error 0
perimeter error 0
area error 0
smoothness error 0
compactness error 0
concavity error 0
concave points error 0
symmetry error 0
fractal dimension error 0
worst radius 0
worst texture 0
worst perimeter 0
worst area 0
worst smoothness 0
worst compactness 0
worst concavity 0
worst concave points 0
worst symmetry 0
worst fractal dimension 0
target 0
dtype: int64
This data has no missing values in any column
Step 4: Exploratory Data Analysis (EDA)#
# find the summary statistics of all the numerical features
df.describe()
| mean radius | mean texture | mean perimeter | mean area | mean smoothness | mean compactness | mean concavity | mean concave points | mean symmetry | mean fractal dimension | ... | worst texture | worst perimeter | worst area | worst smoothness | worst compactness | worst concavity | worst concave points | worst symmetry | worst fractal dimension | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| count | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | ... | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 | 569.000000 |
| mean | 14.127292 | 19.289649 | 91.969033 | 654.889104 | 0.096360 | 0.104341 | 0.088799 | 0.048919 | 0.181162 | 0.062798 | ... | 25.677223 | 107.261213 | 880.583128 | 0.132369 | 0.254265 | 0.272188 | 0.114606 | 0.290076 | 0.083946 | 0.627417 |
| std | 3.524049 | 4.301036 | 24.298981 | 351.914129 | 0.014064 | 0.052813 | 0.079720 | 0.038803 | 0.027414 | 0.007060 | ... | 6.146258 | 33.602542 | 569.356993 | 0.022832 | 0.157336 | 0.208624 | 0.065732 | 0.061867 | 0.018061 | 0.483918 |
| min | 6.981000 | 9.710000 | 43.790000 | 143.500000 | 0.052630 | 0.019380 | 0.000000 | 0.000000 | 0.106000 | 0.049960 | ... | 12.020000 | 50.410000 | 185.200000 | 0.071170 | 0.027290 | 0.000000 | 0.000000 | 0.156500 | 0.055040 | 0.000000 |
| 25% | 11.700000 | 16.170000 | 75.170000 | 420.300000 | 0.086370 | 0.064920 | 0.029560 | 0.020310 | 0.161900 | 0.057700 | ... | 21.080000 | 84.110000 | 515.300000 | 0.116600 | 0.147200 | 0.114500 | 0.064930 | 0.250400 | 0.071460 | 0.000000 |
| 50% | 13.370000 | 18.840000 | 86.240000 | 551.100000 | 0.095870 | 0.092630 | 0.061540 | 0.033500 | 0.179200 | 0.061540 | ... | 25.410000 | 97.660000 | 686.500000 | 0.131300 | 0.211900 | 0.226700 | 0.099930 | 0.282200 | 0.080040 | 1.000000 |
| 75% | 15.780000 | 21.800000 | 104.100000 | 782.700000 | 0.105300 | 0.130400 | 0.130700 | 0.074000 | 0.195700 | 0.066120 | ... | 29.720000 | 125.400000 | 1084.000000 | 0.146000 | 0.339100 | 0.382900 | 0.161400 | 0.317900 | 0.092080 | 1.000000 |
| max | 28.110000 | 39.280000 | 188.500000 | 2501.000000 | 0.163400 | 0.345400 | 0.426800 | 0.201200 | 0.304000 | 0.097440 | ... | 49.540000 | 251.200000 | 4254.000000 | 0.222600 | 1.058000 | 1.252000 | 0.291000 | 0.663800 | 0.207500 | 1.000000 |
8 rows × 31 columns
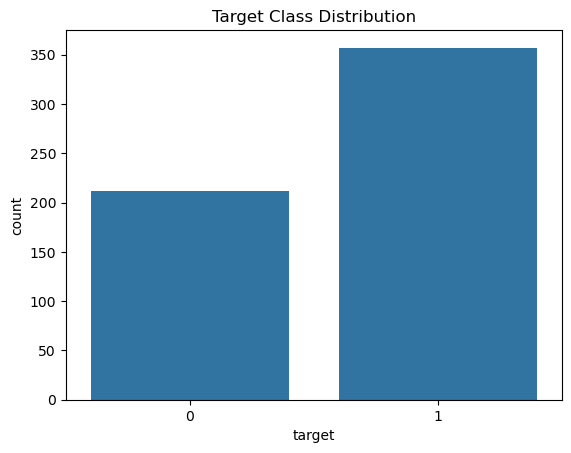
# Plot the number of classes in target variable (column y)
sns.countplot(x='target', data=df)
plt.title('Target Class Distribution')
plt.show()
# plot heatmap of correlation between all the features of dataset df; we are interested in the correlation between the features (X)
plt.figure(figsize=(12, 10))
sns.heatmap(df.corr(), cmap='coolwarm', square=True)
plt.title('Feature Correlation')
plt.show()
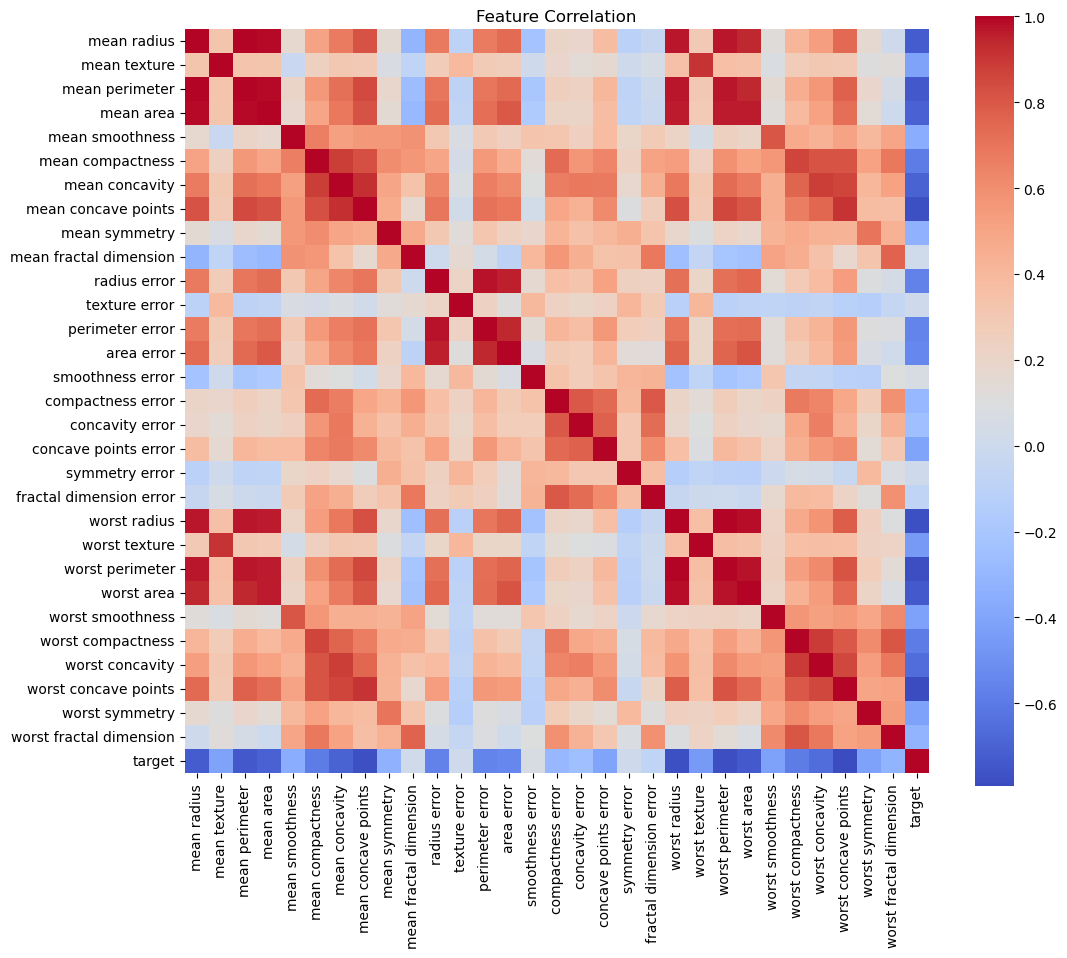
The darker red color shows the high correlation between features. At this point, we may discard one of the two highly correlated features and do further analysis.
Here we check if the two classed are represented approximately equally. If this representation is very skewed towards any one class, we rebalance it using techniques like SMOTE.
Step 5: Split the data randomly into training and testing set#
# Choose all columns of the dataframe except the "target" column as X(input) and "target" column as y(output)
X = df.drop('target', axis=1)
y = df['target']
#Randomly split the data into training an test set in the ratio of 80%(training) and 20% (testing); could be 70/30 or 75/25 also
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=42)
Step 6: Feature Scaling#
Numerical value of feature may be of different magnitudes (for example, max value of “mean smoothness” is 0.163400 wheras max value “mean area” is 2501.00. Hence, we scale the features to transform them to a similar scale (between 0 and 1), so the feature with high numerical values does not dominate the model creation.
Only feature values are scaled, NOT the target values.
scaler = StandardScaler()
X_train_scaled = scaler.fit_transform(X_train)
X_test_scaled = scaler.transform(X_test)
Step 7: Train a Classifier#
For example, Random Forest, Decision Tree, logistic Regression, Naive Bayes, XgBoost, and many more
model = RandomForestClassifier(random_state=42)
model.fit(X_train_scaled, y_train)
RandomForestClassifier(random_state=42)In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook.
On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
RandomForestClassifier(random_state=42)
Step 8: Evaluate the Model#
Some important metrics: precision, recall, accuracy, f1-score
#Calculate the predicted value of y (y_pred) using model
y_pred = model.predict(X_test_scaled)
# Evaluate the model and print evaluation metrics
cm = confusion_matrix(y_test, y_pred)
sns.heatmap(cm, annot=True, fmt="d", cmap="Blues")
plt.xlabel("Predicted")
plt.ylabel("Actual")
plt.title("Confusion Matrix")
plt.show()
print(classification_report(y_test, y_pred))
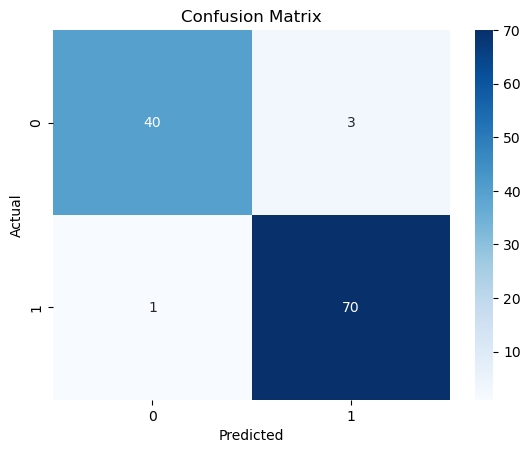
precision recall f1-score support
0 0.98 0.93 0.95 43
1 0.96 0.99 0.97 71
accuracy 0.96 114
macro avg 0.97 0.96 0.96 114
weighted avg 0.97 0.96 0.96 114
The accuracy of the model is 96%
