
This notebook is created by Zhuo Chen under Creative Commons CC BY License
For questions/comments/improvements, email zhuo.chen@ithaka.org or nathan.kelber@ithaka.org
Pandas Basics 3#
Description: This notebook describes how to:
Use
merge()andconcat()to combine multiple dataframesHandle duplicates and get unique values
Understand vectorized operations in Pandas
Use
apply()andwhere()to process the data in a dataframe
This is the third notebook in a series on learning to use Pandas.
Use Case: For Learners (Detailed explanation, not ideal for researchers)
Difficulty: Beginner
Knowledge Required:
Python Basics (Start Python Basics I)
Knowledge Recommended:
Completion Time: 90 minutes
Data Format: csv
Libraries Used: Pandas
Research Pipeline: None
# Import pandas library, `as pd` allows us to shorten typing `pandas` to `pd` when we call pandas
import pandas as pd
Merge and concatenate dataframes#
In this section, we will learn two methods to combine multiple dataframes in Pandas.
merge()#
When dealing with tabular data, we often find ourselves in a situation where we need to merge two or more dataframes as we want a subset of the data from each. Pandas provides a .merge() method that allows us to merge dataframes easily.
The data we use in this section is the unemployment data in Massachusetts from 2023 January to February.
# Get the urls to the files and download the sample files
import urllib.request
from pathlib import Path
# Check if a data folder exists. If not, create it.
data_folder = Path('../data/')
data_folder.mkdir(exist_ok=True)
# Download the files
urls = ['https://ithaka-labs.s3.amazonaws.com/static-files/images/tdm/tdmdocs/Pandas3_unemp_jan.csv',
'https://ithaka-labs.s3.amazonaws.com/static-files/images/tdm/tdmdocs/Pandas3_unemp_feb.csv']
for url in urls:
urllib.request.urlretrieve(url, '../data/'+url.rsplit('/')[-1])
# Success message
print('Sample files ready.')
Sample files ready.
# create a dataframe from the csv files
unemp_jan = pd.read_csv('../data/Pandas3_unemp_jan.csv')
unemp_feb = pd.read_csv('../data/Pandas3_unemp_feb.csv')
# take a look at unemp_jan
unemp_jan
| COUNTY | Jan Unemployment | |
|---|---|---|
| 0 | BARNSTABLE | 7233 |
| 1 | BERKSHIRE | 3078 |
| 2 | BRISTOL | 15952 |
| 3 | DUKES | 696 |
| 4 | ESSEX | 17573 |
| 5 | FRANKLIN | 1506 |
| 6 | HAMPDEN | 11629 |
| 7 | HAMPSHIRE | 3354 |
| 8 | MIDDLESEX | 29445 |
| 9 | NANTUCKET | 949 |
| 10 | NORFOLK | 14193 |
| 11 | PLYMOUTH | 12189 |
| 12 | SUFFOLK | 16410 |
| 13 | WORCESTER | 18944 |
# take a look at unemp_feb
unemp_feb
| COUNTY | Feb Unemployment | |
|---|---|---|
| 0 | BARNSTABLE | 7855 |
| 1 | BERKSHIRE | 3162 |
| 2 | BRISTOL | 17112 |
| 3 | DUKES | 772 |
| 4 | ESSEX | 18486 |
| 5 | FRANKLIN | 1557 |
| 6 | HAMPDEN | 11941 |
| 7 | HAMPSHIRE | 3342 |
| 8 | MIDDLESEX | 29901 |
| 9 | NANTUCKET | 1072 |
| 10 | NORFOLK | 14356 |
| 11 | PLYMOUTH | 12798 |
| 12 | SUFFOLK | 16318 |
| 13 | WORCESTER | 20135 |
The two dataframes have a column in common, the COUNTY column. This column contains the names of the 14 counties in the state of Massachusetts.
inner join#
# merge the two dfs
unemp_jan.merge(unemp_feb, on='COUNTY')
| COUNTY | Jan Unemployment | Feb Unemployment | |
|---|---|---|---|
| 0 | BARNSTABLE | 7233 | 7855 |
| 1 | BERKSHIRE | 3078 | 3162 |
| 2 | BRISTOL | 15952 | 17112 |
| 3 | DUKES | 696 | 772 |
| 4 | ESSEX | 17573 | 18486 |
| 5 | FRANKLIN | 1506 | 1557 |
| 6 | HAMPDEN | 11629 | 11941 |
| 7 | HAMPSHIRE | 3354 | 3342 |
| 8 | MIDDLESEX | 29445 | 29901 |
| 9 | NANTUCKET | 949 | 1072 |
| 10 | NORFOLK | 14193 | 14356 |
| 11 | PLYMOUTH | 12189 | 12798 |
| 12 | SUFFOLK | 16410 | 16318 |
| 13 | WORCESTER | 18944 | 20135 |
By convention, we’ll call the dataframe on which we call the .merge() method the left dataframe. We’ll call the dataframe passed into the .merge() method the right dataframe.
As you can see, the parameter ‘on’ specifies the key column we use to merge the two dataframes. By default, the type of merge performed is ‘inner’, which uses the intersection of the keys from the key column. You can explicitly specify the type of merge by setting the ‘how’ parameter.
# set the parameter 'how' to specify the merge type
unemp_jan.merge(unemp_feb, how='inner', on='COUNTY')
| COUNTY | Jan Unemployment | Feb Unemployment | |
|---|---|---|---|
| 0 | BARNSTABLE | 7233 | 7855 |
| 1 | BERKSHIRE | 3078 | 3162 |
| 2 | BRISTOL | 15952 | 17112 |
| 3 | DUKES | 696 | 772 |
| 4 | ESSEX | 17573 | 18486 |
| 5 | FRANKLIN | 1506 | 1557 |
| 6 | HAMPDEN | 11629 | 11941 |
| 7 | HAMPSHIRE | 3354 | 3342 |
| 8 | MIDDLESEX | 29445 | 29901 |
| 9 | NANTUCKET | 949 | 1072 |
| 10 | NORFOLK | 14193 | 14356 |
| 11 | PLYMOUTH | 12189 | 12798 |
| 12 | SUFFOLK | 16410 | 16318 |
| 13 | WORCESTER | 18944 | 20135 |
In an inner join, you’ll lose the rows that do not have a matching key in the two dataframes. In other words, only rows from the two dataframes that have a matching value in the key column will appear after the merge.
# see what happens when there are non-matching keys
a = unemp_jan.drop(0)
b = unemp_feb.drop(13)
a.merge(b, on='COUNTY')
| COUNTY | Jan Unemployment | Feb Unemployment | |
|---|---|---|---|
| 0 | BERKSHIRE | 3078 | 3162 |
| 1 | BRISTOL | 15952 | 17112 |
| 2 | DUKES | 696 | 772 |
| 3 | ESSEX | 17573 | 18486 |
| 4 | FRANKLIN | 1506 | 1557 |
| 5 | HAMPDEN | 11629 | 11941 |
| 6 | HAMPSHIRE | 3354 | 3342 |
| 7 | MIDDLESEX | 29445 | 29901 |
| 8 | NANTUCKET | 949 | 1072 |
| 9 | NORFOLK | 14193 | 14356 |
| 10 | PLYMOUTH | 12189 | 12798 |
| 11 | SUFFOLK | 16410 | 16318 |
As you can see, we have lost the data for Barnstable and Worcester after the merge. This is because the former has been dropped from the left dataframe and the latter has been dropped from the right dataframe.
outer join#
There are other types of merge, of course. Instead of using the intersection of keys from two dataframes, we can use the union of keys from two dataframes. The type of merge in this case is ‘outer’.
# change the merge type and see what happens
a = unemp_jan.drop(0)
b = unemp_feb.drop(13)
a.merge(b, how='outer', on='COUNTY')
| COUNTY | Jan Unemployment | Feb Unemployment | |
|---|---|---|---|
| 0 | BARNSTABLE | NaN | 7855.0 |
| 1 | BERKSHIRE | 3078.0 | 3162.0 |
| 2 | BRISTOL | 15952.0 | 17112.0 |
| 3 | DUKES | 696.0 | 772.0 |
| 4 | ESSEX | 17573.0 | 18486.0 |
| 5 | FRANKLIN | 1506.0 | 1557.0 |
| 6 | HAMPDEN | 11629.0 | 11941.0 |
| 7 | HAMPSHIRE | 3354.0 | 3342.0 |
| 8 | MIDDLESEX | 29445.0 | 29901.0 |
| 9 | NANTUCKET | 949.0 | 1072.0 |
| 10 | NORFOLK | 14193.0 | 14356.0 |
| 11 | PLYMOUTH | 12189.0 | 12798.0 |
| 12 | SUFFOLK | 16410.0 | 16318.0 |
| 13 | WORCESTER | 18944.0 | NaN |
As you can see, the union of the values in the ‘COUNTY’ column from the two dataframes is used to merge the two. For keys that only exist in one dataframe, unmatched columns will be filled in with NaN.
left outer join#
Still another type of merge is to use the keys from the left dataframe in the merge. The values in the key column from the left dataframe will be used to merge the two dataframes.
# Use the keys from the left df in the merge
a = unemp_jan.drop(0)
b = unemp_feb.drop(13)
a.merge(b, how='left', on='COUNTY')
| COUNTY | Jan Unemployment | Feb Unemployment | |
|---|---|---|---|
| 0 | BERKSHIRE | 3078 | 3162.0 |
| 1 | BRISTOL | 15952 | 17112.0 |
| 2 | DUKES | 696 | 772.0 |
| 3 | ESSEX | 17573 | 18486.0 |
| 4 | FRANKLIN | 1506 | 1557.0 |
| 5 | HAMPDEN | 11629 | 11941.0 |
| 6 | HAMPSHIRE | 3354 | 3342.0 |
| 7 | MIDDLESEX | 29445 | 29901.0 |
| 8 | NANTUCKET | 949 | 1072.0 |
| 9 | NORFOLK | 14193 | 14356.0 |
| 10 | PLYMOUTH | 12189 | 12798.0 |
| 11 | SUFFOLK | 16410 | 16318.0 |
| 12 | WORCESTER | 18944 | NaN |
# see what happens if right df has a non-matching key
unemp_jan.merge(unemp_feb.drop(0), how='left', on='COUNTY')
| COUNTY | Jan Unemployment | Feb Unemployment | |
|---|---|---|---|
| 0 | BARNSTABLE | 7233 | NaN |
| 1 | BERKSHIRE | 3078 | 3162.0 |
| 2 | BRISTOL | 15952 | 17112.0 |
| 3 | DUKES | 696 | 772.0 |
| 4 | ESSEX | 17573 | 18486.0 |
| 5 | FRANKLIN | 1506 | 1557.0 |
| 6 | HAMPDEN | 11629 | 11941.0 |
| 7 | HAMPSHIRE | 3354 | 3342.0 |
| 8 | MIDDLESEX | 29445 | 29901.0 |
| 9 | NANTUCKET | 949 | 1072.0 |
| 10 | NORFOLK | 14193 | 14356.0 |
| 11 | PLYMOUTH | 12189 | 12798.0 |
| 12 | SUFFOLK | 16410 | 16318.0 |
| 13 | WORCESTER | 18944 | 20135.0 |
right outer join#
Or, you can use the keys from the right dataframe in the merge.
# Use the keys from the right df in the merge
unemp_jan.merge(unemp_feb.drop(0), how='right', on='COUNTY')
| COUNTY | Jan Unemployment | Feb Unemployment | |
|---|---|---|---|
| 0 | BERKSHIRE | 3078 | 3162 |
| 1 | BRISTOL | 15952 | 17112 |
| 2 | DUKES | 696 | 772 |
| 3 | ESSEX | 17573 | 18486 |
| 4 | FRANKLIN | 1506 | 1557 |
| 5 | HAMPDEN | 11629 | 11941 |
| 6 | HAMPSHIRE | 3354 | 3342 |
| 7 | MIDDLESEX | 29445 | 29901 |
| 8 | NANTUCKET | 949 | 1072 |
| 9 | NORFOLK | 14193 | 14356 |
| 10 | PLYMOUTH | 12189 | 12798 |
| 11 | SUFFOLK | 16410 | 16318 |
| 12 | WORCESTER | 18944 | 20135 |
# see what happens if left df has a non-matching key
unemp_jan.drop(0).merge(unemp_feb, how='right', on='COUNTY')
| COUNTY | Jan Unemployment | Feb Unemployment | |
|---|---|---|---|
| 0 | BARNSTABLE | NaN | 7855 |
| 1 | BERKSHIRE | 3078.0 | 3162 |
| 2 | BRISTOL | 15952.0 | 17112 |
| 3 | DUKES | 696.0 | 772 |
| 4 | ESSEX | 17573.0 | 18486 |
| 5 | FRANKLIN | 1506.0 | 1557 |
| 6 | HAMPDEN | 11629.0 | 11941 |
| 7 | HAMPSHIRE | 3354.0 | 3342 |
| 8 | MIDDLESEX | 29445.0 | 29901 |
| 9 | NANTUCKET | 949.0 | 1072 |
| 10 | NORFOLK | 14193.0 | 14356 |
| 11 | PLYMOUTH | 12189.0 | 12798 |
| 12 | SUFFOLK | 16410.0 | 16318 |
| 13 | WORCESTER | 18944.0 | 20135 |
The suffix parameter#
By default, Pandas does not allow duplicate column names. Therefore, if the two dataframes have a column other than the key column that has the same name, Pandas automatically uses suffixes to distinguish them after the merge.
# create a df with the unemployment data of MA in Feb of 2022
unemp_feb_22 = pd.DataFrame({'COUNTY':
['BARNSTABLE',
'BERKSHIRE',
'BRISTOL',
'DUKES',
'ESSEX',
'FRANKLIN',
'HAMPDEN',
'HAMPSHIRE',
'MIDDLESEX',
'NANTUCKET',
'NORFOLK',
'PLYMOUTH',
'SUFFOLK',
'WORCESTER'],
'Feb Unemployment':
[7952,
3366,
17777,
804,
19134,
1653,
12830,
3305,
29335,
1071,
14045,
14072,
17043,
20000]})
unemp_feb_22
| COUNTY | Feb Unemployment | |
|---|---|---|
| 0 | BARNSTABLE | 7952 |
| 1 | BERKSHIRE | 3366 |
| 2 | BRISTOL | 17777 |
| 3 | DUKES | 804 |
| 4 | ESSEX | 19134 |
| 5 | FRANKLIN | 1653 |
| 6 | HAMPDEN | 12830 |
| 7 | HAMPSHIRE | 3305 |
| 8 | MIDDLESEX | 29335 |
| 9 | NANTUCKET | 1071 |
| 10 | NORFOLK | 14045 |
| 11 | PLYMOUTH | 14072 |
| 12 | SUFFOLK | 17043 |
| 13 | WORCESTER | 20000 |
# merge the two and see what happens
unemp_feb.merge(unemp_feb_22, on='COUNTY')
| COUNTY | Feb Unemployment_x | Feb Unemployment_y | |
|---|---|---|---|
| 0 | BARNSTABLE | 7855 | 7952 |
| 1 | BERKSHIRE | 3162 | 3366 |
| 2 | BRISTOL | 17112 | 17777 |
| 3 | DUKES | 772 | 804 |
| 4 | ESSEX | 18486 | 19134 |
| 5 | FRANKLIN | 1557 | 1653 |
| 6 | HAMPDEN | 11941 | 12830 |
| 7 | HAMPSHIRE | 3342 | 3305 |
| 8 | MIDDLESEX | 29901 | 29335 |
| 9 | NANTUCKET | 1072 | 1071 |
| 10 | NORFOLK | 14356 | 14045 |
| 11 | PLYMOUTH | 12798 | 14072 |
| 12 | SUFFOLK | 16318 | 17043 |
| 13 | WORCESTER | 20135 | 20000 |
The suffix ‘_x’ indicates that the column is from the left dataframe. The suffix ‘_y’ indicates that the column is from the right dataframe. You can make the suffixes more descriptive by setting the the value of the parameter ‘suffixes’.
# make the suffixes more descriptive
unemp_feb.merge(unemp_feb_22, on='COUNTY', suffixes=[' 2023', ' 2022'])
| COUNTY | Feb Unemployment 2023 | Feb Unemployment 2022 | |
|---|---|---|---|
| 0 | BARNSTABLE | 7855 | 7952 |
| 1 | BERKSHIRE | 3162 | 3366 |
| 2 | BRISTOL | 17112 | 17777 |
| 3 | DUKES | 772 | 804 |
| 4 | ESSEX | 18486 | 19134 |
| 5 | FRANKLIN | 1557 | 1653 |
| 6 | HAMPDEN | 11941 | 12830 |
| 7 | HAMPSHIRE | 3342 | 3305 |
| 8 | MIDDLESEX | 29901 | 29335 |
| 9 | NANTUCKET | 1072 | 1071 |
| 10 | NORFOLK | 14356 | 14045 |
| 11 | PLYMOUTH | 12798 | 14072 |
| 12 | SUFFOLK | 16318 | 17043 |
| 13 | WORCESTER | 20135 | 20000 |
concat()#
The .concat() method stitches two dataframes together along the row axis or the column axis. It is often used to combine two datasets to form a larger one for further processing.
By default, the concat() method concatenates multiple dataframes along the row axis.
# concatenate unemp_feb and unemp_feb_22
pd.concat([unemp_feb, unemp_feb_22])
| COUNTY | Feb Unemployment | |
|---|---|---|
| 0 | BARNSTABLE | 7855 |
| 1 | BERKSHIRE | 3162 |
| 2 | BRISTOL | 17112 |
| 3 | DUKES | 772 |
| 4 | ESSEX | 18486 |
| 5 | FRANKLIN | 1557 |
| 6 | HAMPDEN | 11941 |
| 7 | HAMPSHIRE | 3342 |
| 8 | MIDDLESEX | 29901 |
| 9 | NANTUCKET | 1072 |
| 10 | NORFOLK | 14356 |
| 11 | PLYMOUTH | 12798 |
| 12 | SUFFOLK | 16318 |
| 13 | WORCESTER | 20135 |
| 0 | BARNSTABLE | 7952 |
| 1 | BERKSHIRE | 3366 |
| 2 | BRISTOL | 17777 |
| 3 | DUKES | 804 |
| 4 | ESSEX | 19134 |
| 5 | FRANKLIN | 1653 |
| 6 | HAMPDEN | 12830 |
| 7 | HAMPSHIRE | 3305 |
| 8 | MIDDLESEX | 29335 |
| 9 | NANTUCKET | 1071 |
| 10 | NORFOLK | 14045 |
| 11 | PLYMOUTH | 14072 |
| 12 | SUFFOLK | 17043 |
| 13 | WORCESTER | 20000 |
You can see that the indexes from the original dataframes are preserved after the concatenation. If you would like to reset the index after concatenation, you can set the parameter ignore_index to True.
# set ignore_index to False
pd.concat([unemp_feb, unemp_feb_22], ignore_index=True)
| COUNTY | Feb Unemployment | |
|---|---|---|
| 0 | BARNSTABLE | 7855 |
| 1 | BERKSHIRE | 3162 |
| 2 | BRISTOL | 17112 |
| 3 | DUKES | 772 |
| 4 | ESSEX | 18486 |
| 5 | FRANKLIN | 1557 |
| 6 | HAMPDEN | 11941 |
| 7 | HAMPSHIRE | 3342 |
| 8 | MIDDLESEX | 29901 |
| 9 | NANTUCKET | 1072 |
| 10 | NORFOLK | 14356 |
| 11 | PLYMOUTH | 12798 |
| 12 | SUFFOLK | 16318 |
| 13 | WORCESTER | 20135 |
| 14 | BARNSTABLE | 7952 |
| 15 | BERKSHIRE | 3366 |
| 16 | BRISTOL | 17777 |
| 17 | DUKES | 804 |
| 18 | ESSEX | 19134 |
| 19 | FRANKLIN | 1653 |
| 20 | HAMPDEN | 12830 |
| 21 | HAMPSHIRE | 3305 |
| 22 | MIDDLESEX | 29335 |
| 23 | NANTUCKET | 1071 |
| 24 | NORFOLK | 14045 |
| 25 | PLYMOUTH | 14072 |
| 26 | SUFFOLK | 17043 |
| 27 | WORCESTER | 20000 |
The two dataframes unemp_feb and unemp_feb_22 have the same columns. Both have a column COUNTY and a column Feb Unemployment. What if we have two dataframes that have non-matching columns? For example, the dataframe unemp_jan has a COUNTY column and a Jan Unemployment column. If we concatenate unemp_feb and unemp_jan, what will happen?
# concatenate two dfs that have non-matching columns
pd.concat([unemp_feb, unemp_jan])
| COUNTY | Feb Unemployment | Jan Unemployment | |
|---|---|---|---|
| 0 | BARNSTABLE | 7855.0 | NaN |
| 1 | BERKSHIRE | 3162.0 | NaN |
| 2 | BRISTOL | 17112.0 | NaN |
| 3 | DUKES | 772.0 | NaN |
| 4 | ESSEX | 18486.0 | NaN |
| 5 | FRANKLIN | 1557.0 | NaN |
| 6 | HAMPDEN | 11941.0 | NaN |
| 7 | HAMPSHIRE | 3342.0 | NaN |
| 8 | MIDDLESEX | 29901.0 | NaN |
| 9 | NANTUCKET | 1072.0 | NaN |
| 10 | NORFOLK | 14356.0 | NaN |
| 11 | PLYMOUTH | 12798.0 | NaN |
| 12 | SUFFOLK | 16318.0 | NaN |
| 13 | WORCESTER | 20135.0 | NaN |
| 0 | BARNSTABLE | NaN | 7233.0 |
| 1 | BERKSHIRE | NaN | 3078.0 |
| 2 | BRISTOL | NaN | 15952.0 |
| 3 | DUKES | NaN | 696.0 |
| 4 | ESSEX | NaN | 17573.0 |
| 5 | FRANKLIN | NaN | 1506.0 |
| 6 | HAMPDEN | NaN | 11629.0 |
| 7 | HAMPSHIRE | NaN | 3354.0 |
| 8 | MIDDLESEX | NaN | 29445.0 |
| 9 | NANTUCKET | NaN | 949.0 |
| 10 | NORFOLK | NaN | 14193.0 |
| 11 | PLYMOUTH | NaN | 12189.0 |
| 12 | SUFFOLK | NaN | 16410.0 |
| 13 | WORCESTER | NaN | 18944.0 |
If you would like to concatenate two dataframes along the column axis, just set the value of the ‘axis’ parameter to 1.
# concatenate along the column axis
pd.concat([unemp_jan.set_index('COUNTY'), unemp_feb.set_index('COUNTY')], axis=1)
| Jan Unemployment | Feb Unemployment | |
|---|---|---|
| COUNTY | ||
| BARNSTABLE | 7233 | 7855 |
| BERKSHIRE | 3078 | 3162 |
| BRISTOL | 15952 | 17112 |
| DUKES | 696 | 772 |
| ESSEX | 17573 | 18486 |
| FRANKLIN | 1506 | 1557 |
| HAMPDEN | 11629 | 11941 |
| HAMPSHIRE | 3354 | 3342 |
| MIDDLESEX | 29445 | 29901 |
| NANTUCKET | 949 | 1072 |
| NORFOLK | 14193 | 14356 |
| PLYMOUTH | 12189 | 12798 |
| SUFFOLK | 16410 | 16318 |
| WORCESTER | 18944 | 20135 |
Duplicates and unique values#
After we combine several dataframes into a big one, a common practice is to remove the duplicates in the big dataframe.
drop_duplicates()#
There is a handy method in Pandas that can remove duplicates, i.e. drop_duplicates().
# make a df with duplicates
rate = pd.DataFrame([['Pandas', 'Morning', 7],
['Pandas', 'Morning', 7],
['Pandas', 'Evening', 8],
['PythonBasics','Morning', 9]],
columns=['Course', 'Session', 'Rating'])
rate
| Course | Session | Rating | |
|---|---|---|---|
| 0 | Pandas | Morning | 7 |
| 1 | Pandas | Morning | 7 |
| 2 | Pandas | Evening | 8 |
| 3 | PythonBasics | Morning | 9 |
By default, .drop_duplicates() considers all columns when removing duplicates.
# drop duplicates
rate.drop_duplicates()
| Course | Session | Rating | |
|---|---|---|---|
| 0 | Pandas | Morning | 7 |
| 2 | Pandas | Evening | 8 |
| 3 | PythonBasics | Morning | 9 |
You can specify the column(s) to look for duplicates by setting the subset parameter.
# set the 'subset' parameter
rate.drop_duplicates(subset=['Course'])
| Course | Session | Rating | |
|---|---|---|---|
| 0 | Pandas | Morning | 7 |
| 3 | PythonBasics | Morning | 9 |
By default, the first occurrence of the duplicates will be kept. If you would like to keep the last occurrence, you can set the keep parameter to last.
# set the 'keep' parameter to 'last'
rate.drop_duplicates(subset=['Course'], keep='last')
| Course | Session | Rating | |
|---|---|---|---|
| 2 | Pandas | Evening | 8 |
| 3 | PythonBasics | Morning | 9 |
To drop all duplicates, you can set the keep parameter to False.
# drop all duplicates
rate.drop_duplicates(subset=['Course'], keep=False)
| Course | Session | Rating | |
|---|---|---|---|
| 3 | PythonBasics | Morning | 9 |
After you remove all duplicates from a column, the values left in that column are unique.
There is another useful method to find the unique values in a certain column in a dataframe is the .unique() method
# Get the unique courses in df rate
rate['Course'].unique()
array(['Pandas', 'PythonBasics'], dtype=object)
Coding Challenge! < / >
In this coding challenge, we’ll work with the data on the 2021 and 2022 Boston Marathon.
# Get the urls to the files and download the files
import urllib.request
urls = ['https://ithaka-labs.s3.amazonaws.com/static-files/images/tdm/tdmdocs/DataViz3_BostonMarathon2021.csv',
'https://ithaka-labs.s3.amazonaws.com/static-files/images/tdm/tdmdocs/DataViz3_BostonMarathon2022.csv']
for url in urls:
urllib.request.urlretrieve(url, '../data/' + url.rsplit('/')[-1][9:])
# Success message
print('Sample files ready.')
Sample files ready.
# create dfs out of the files
bm_21 = pd.read_csv('../data/BostonMarathon2021.csv')
bm_22 = pd.read_csv('../data/BostonMarathon2022.csv')
# explore bm_21
bm_21.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 15386 entries, 0 to 15385
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 BibNumber 15386 non-null int64
1 FullName 15386 non-null object
2 SortName 15386 non-null object
3 AgeOnRaceDay 15386 non-null int64
4 Gender 15386 non-null object
5 City 15357 non-null object
6 StateAbbrev 14651 non-null object
7 StateName 14651 non-null object
8 Zip 15308 non-null object
9 CountryOfResAbbrev 15386 non-null object
10 CountryOfResName 15386 non-null object
11 CountryOfCtzAbbrev 15386 non-null object
12 CountryOfCtzName 15386 non-null object
13 OfficialTime 15386 non-null object
14 RankOverall 15386 non-null int64
15 RankOverGender 15386 non-null int64
16 RankOverDivision 15386 non-null int64
17 EventGroup 15386 non-null object
18 SubGroupLabel 27 non-null object
19 SubGroup 27 non-null object
dtypes: int64(5), object(15)
memory usage: 2.3+ MB
# explore bm_22
bm_22.info()
<class 'pandas.core.frame.DataFrame'>
RangeIndex: 24834 entries, 0 to 24833
Data columns (total 20 columns):
# Column Non-Null Count Dtype
--- ------ -------------- -----
0 BibNumber 24834 non-null int64
1 FullName 24834 non-null object
2 SortName 24834 non-null object
3 AgeOnRaceDay 24834 non-null int64
4 Gender 24834 non-null object
5 City 24820 non-null object
6 StateAbbrev 20127 non-null object
7 StateName 20127 non-null object
8 Zip 24620 non-null object
9 CountryOfResAbbrev 24833 non-null object
10 CountryOfResName 24833 non-null object
11 CountryOfCtzAbbrev 24806 non-null object
12 CountryOfCtzName 24806 non-null object
13 OfficialTime 24834 non-null object
14 RankOverall 24834 non-null int64
15 RankOverGender 24834 non-null int64
16 RankOverDivision 24834 non-null int64
17 EventGroup 24834 non-null object
18 SubGroupLabel 31 non-null object
19 SubGroup 31 non-null object
dtypes: int64(5), object(15)
memory usage: 3.8+ MB
Using what you have learned so far, can you find out which countries have runners in 2021 Boston Marathon but not 2022 Boston Marathon? Which countries have runners in 2022 Boston Marathon but not 2021 Boston Marathon? Let’s get the countries using the CountryOfResName column in this exercise.
# find out which countries have runners in 2021 but not in 2022
# find out which countries have runners in 2022 but not in 2021
Vectorized operations in Pandas#
As you have seen so far, a lot of the methods in Pandas can work on the values in a row or a column in a batch. For example, if you call .isna() on a column, it checks, for each value in that column, whether it is NaN or not. This kind of operation where values are taken and operated on in a batch is called vectorized operations in Pandas.
# a simple example of vectorized operation
df = pd.DataFrame({'number':[1,2,3,4],
'year': [2020, 2021, 2022, 2023]})
# grab the 'number' column and add 2 to it
df['number'] + 2
0 3
1 4
2 5
3 6
Name: number, dtype: int64
As you can see, the addition is applied element-wise to the integers in the number column. How do we understand this operation? Why don’t we need to write a for loop to iterate over the values in the number column and add 2 to each of them?
Actually, this vectorized addition can be broken down into two steps.
First of all, the addend 2 undergoes a process called broadcasting. It is stretched into a vector of the same shape as the
numbercolumn.
Second, we add the integers in the
numbercolumn and the vector of 2s in the same way that we add two vectors in mathematics. We have vectorized the addition in this case.
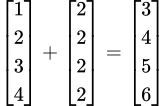
When we add two vectors, the two numbers in the corresponding positions in the two vectors are added together.
The importance of shape#
A prerequisite for applying an operation to two vectors is to make sure that they have compatible shapes. In the previous example, we are able to add 2 to the number column in a vectorized way because the value 2 can be broadcast into a vector whose shape is compatible with the number column.
# applying an operation to two vectors of incompatible shapes
# throws an error
df['number'] + [1, 2]
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[36], line 3
1 # applying an operation to two vectors of incompatible shapes
2 # throws an error
----> 3 df['number'] + [1, 2]
File /opt/homebrew/lib/python3.11/site-packages/pandas/core/ops/common.py:76, in _unpack_zerodim_and_defer.<locals>.new_method(self, other)
72 return NotImplemented
74 other = item_from_zerodim(other)
---> 76 return method(self, other)
File /opt/homebrew/lib/python3.11/site-packages/pandas/core/arraylike.py:186, in OpsMixin.__add__(self, other)
98 @unpack_zerodim_and_defer("__add__")
99 def __add__(self, other):
100 """
101 Get Addition of DataFrame and other, column-wise.
102
(...) 184 moose 3.0 NaN
185 """
--> 186 return self._arith_method(other, operator.add)
File /opt/homebrew/lib/python3.11/site-packages/pandas/core/series.py:6146, in Series._arith_method(self, other, op)
6144 def _arith_method(self, other, op):
6145 self, other = self._align_for_op(other)
-> 6146 return base.IndexOpsMixin._arith_method(self, other, op)
File /opt/homebrew/lib/python3.11/site-packages/pandas/core/base.py:1391, in IndexOpsMixin._arith_method(self, other, op)
1388 rvalues = np.arange(rvalues.start, rvalues.stop, rvalues.step)
1390 with np.errstate(all="ignore"):
-> 1391 result = ops.arithmetic_op(lvalues, rvalues, op)
1393 return self._construct_result(result, name=res_name)
File /opt/homebrew/lib/python3.11/site-packages/pandas/core/ops/array_ops.py:283, in arithmetic_op(left, right, op)
279 _bool_arith_check(op, left, right) # type: ignore[arg-type]
281 # error: Argument 1 to "_na_arithmetic_op" has incompatible type
282 # "Union[ExtensionArray, ndarray[Any, Any]]"; expected "ndarray[Any, Any]"
--> 283 res_values = _na_arithmetic_op(left, right, op) # type: ignore[arg-type]
285 return res_values
File /opt/homebrew/lib/python3.11/site-packages/pandas/core/ops/array_ops.py:218, in _na_arithmetic_op(left, right, op, is_cmp)
215 func = partial(expressions.evaluate, op)
217 try:
--> 218 result = func(left, right)
219 except TypeError:
220 if not is_cmp and (
221 left.dtype == object or getattr(right, "dtype", None) == object
222 ):
(...) 225 # Don't do this for comparisons, as that will handle complex numbers
226 # incorrectly, see GH#32047
File /opt/homebrew/lib/python3.11/site-packages/pandas/core/computation/expressions.py:242, in evaluate(op, a, b, use_numexpr)
239 if op_str is not None:
240 if use_numexpr:
241 # error: "None" not callable
--> 242 return _evaluate(op, op_str, a, b) # type: ignore[misc]
243 return _evaluate_standard(op, op_str, a, b)
File /opt/homebrew/lib/python3.11/site-packages/pandas/core/computation/expressions.py:73, in _evaluate_standard(op, op_str, a, b)
71 if _TEST_MODE:
72 _store_test_result(False)
---> 73 return op(a, b)
ValueError: operands could not be broadcast together with shapes (4,) (2,)
Note that because the vectorized operations apply element-wise, we do not need to write a for loop to iterate over the values in a column and repeat the desired operation to each value.
Whenever you are tempted to write a for loop when processing data in Pandas, take a pause and think about whether there is a vectorized way to do it. In most cases, vectorized operations are faster and more efficient.
# create a df with 1000 rows of random ints between 1 and 1000
import numpy as np
df_test = pd.DataFrame({'a':np.random.randint(1,1000, 1000),
'b':np.random.randint(1,1000, 1000)})
Let’s use a for loop to add two columns and get the execution time.
%%timeit -n 10
for index, row in df_test.iterrows():
row['sum'] = row['a'] + row['b']
110 ms ± 2.06 ms per loop (mean ± std. dev. of 7 runs, 10 loops each)
Let’s add two columns in a vectorized way and get the execution time.
%%timeit -n 10
df_test['sum'] = df_test['a'] + df_test['b']
92.3 μs ± 35.4 μs per loop (mean ± std. dev. of 7 runs, 10 loops each)
Some more examples of vectorized operations#
With numerical data, it is really convenient to use vectorized operations.
# Get the unemployment rate in MA in feb 2023
unemp_feb['Feb Labor Force'] = [108646,
61320,
300028,
8403,
424616,
40085,
225735,
90085,
914953,
6837,
395358,
285373,
455084,
446151]
unemp_feb['Feb Unemployment Rate'] = (unemp_feb['Feb Unemployment']/
unemp_feb['Feb Labor Force'])
unemp_feb
| COUNTY | Feb Unemployment | Feb Labor Force | Feb Unemployment Rate | |
|---|---|---|---|---|
| 0 | BARNSTABLE | 7855 | 108646 | 0.072299 |
| 1 | BERKSHIRE | 3162 | 61320 | 0.051566 |
| 2 | BRISTOL | 17112 | 300028 | 0.057035 |
| 3 | DUKES | 772 | 8403 | 0.091872 |
| 4 | ESSEX | 18486 | 424616 | 0.043536 |
| 5 | FRANKLIN | 1557 | 40085 | 0.038842 |
| 6 | HAMPDEN | 11941 | 225735 | 0.052898 |
| 7 | HAMPSHIRE | 3342 | 90085 | 0.037098 |
| 8 | MIDDLESEX | 29901 | 914953 | 0.032680 |
| 9 | NANTUCKET | 1072 | 6837 | 0.156794 |
| 10 | NORFOLK | 14356 | 395358 | 0.036311 |
| 11 | PLYMOUTH | 12798 | 285373 | 0.044847 |
| 12 | SUFFOLK | 16318 | 455084 | 0.035857 |
| 13 | WORCESTER | 20135 | 446151 | 0.045130 |
You have learned a bunch of string methods in Python Basics. Almost all Python’s built-in string methods have counterparts in Pandas’s vectorized string methods, e.g., upper(), lower(), isalpha(), startswith(), split(), so on and so forth.
# make all county names in lower case
unemp_jan['COUNTY'].str.lower()
0 barnstable
1 berkshire
2 bristol
3 dukes
4 essex
5 franklin
6 hampden
7 hampshire
8 middlesex
9 nantucket
10 norfolk
11 plymouth
12 suffolk
13 worcester
Name: COUNTY, dtype: object
Note that we will first use .str to access the values in a column as strings and then call a certain method. Let’s see an example of another method .split().
# create a df with some full names
full_name = bm_21['FullName'].copy().loc[:5]
full_name
0 Benson Kipruto
1 Lemi Berhanu
2 Jemal Yimer
3 Tsedat Ayana
4 Leonard Barsoton
5 Bayelign Teshager
Name: FullName, dtype: object
# Split the names into first names and last names
full_name.str.split()
0 [Benson, Kipruto]
1 [Lemi, Berhanu]
2 [Jemal, Yimer]
3 [Tsedat, Ayana]
4 [Leonard, Barsoton]
5 [Bayelign, Teshager]
Name: FullName, dtype: object
With strings, a common operation is to search for a certain substring in them. You have seen an example in Pandas basics 2 where we extract all failed banks whose name contains the word ‘Community’.
# a reminder of the contains() method
full_name.loc[full_name.str.contains('Leo')]
4 Leonard Barsoton
Name: FullName, dtype: object
Another common operation with strings is to find out all strings that match a certain pattern. There are some string methods that accept regular expressions which can be used to specify patterns.
# search strings of a certain pattern
full_name.str.extract('(^L.*[nu]$)')
| 0 | |
|---|---|
| 0 | NaN |
| 1 | Lemi Berhanu |
| 2 | NaN |
| 3 | NaN |
| 4 | Leonard Barsoton |
| 5 | NaN |
If you are interested in learning more about regular expressions, check out this constellate notebook.
apply() and where()#
There are a lot of methods in Pandas used for data processing. In this section, we focus our attention on two: apply() and where().
The .apply() method allows you to apply a self-defined function to rows and columns in a dataframe.
Suppose you have a dataframe that contains the words produced by a baby each day.
# use apply to process data
vocab = pd.DataFrame([[{'duck', 'dog'}, {'dog', 'bird'}],
[{'hippo', 'sky'}, {'sky', 'cloud'}]],
columns=['day1', 'day2'],
index=['Alex', 'Ben'])
vocab
| day1 | day2 | |
|---|---|---|
| Alex | {duck, dog} | {bird, dog} |
| Ben | {sky, hippo} | {cloud, sky} |
Now, suppose for each baby, you would like to extract the words that appear in both days so that you know which words were consistently produced them.
### use apply to get the intersection between sets in each row
# define a function that takes a row and returns the desired intersection
def get_common_words(r):
return r['day1'] & r['day2']
# pass the function to .apply()
vocab.apply(get_common_words, axis=1)
Alex {dog}
Ben {sky}
dtype: object
If you are more comfortable with the lambda function, you can pass a lambda function directly to the .apply() method.
# pass a lambda function to apply()
vocab.apply(lambda r: r['day1'] & r['day2'], axis=1)
Alex {dog}
Ben {sky}
dtype: object
The lambda function may look a little bit complicated to read. A good way to read this function is to break it down into two parts. What goes before the colon is the input to this function; what goes after the colon is the output of this function.
The .where() method allows you to manipulate the data using the if-else logic. The syntax of the .where() method is .where(condition, other). The values fulfilling the condition will be kept and the values that do not fulfill the condition are changed to the value specified by ‘other’.
Suppose you are a middle school teacher. You have a report of the grades for the most English test.
# create a df containing English grades
eng = pd.DataFrame({'grade': [90, 88, 70, 55]},
index=['Alice', 'Becky', 'Cindy', 'Dave'])
eng
| grade | |
|---|---|
| Alice | 90 |
| Becky | 88 |
| Cindy | 70 |
| Dave | 55 |
And you would like to change any grade below 60 to ‘F’.
# use .where() to change any grade below 60 to 'F'
eng.where(eng['grade']>=60, 'F')
| grade | |
|---|---|
| Alice | 90 |
| Becky | 88 |
| Cindy | 70 |
| Dave | F |
You can do the same thing using the .apply() method. However, the if-else logic will need to be written into the function passed into .apply().
# use apply to change the grades below 60 to 'F'
eng['grade'].apply(lambda x: 'F' if x < 60 else x)
Alice 90
Becky 88
Cindy 70
Dave F
Name: grade, dtype: object
We have seen how to apply the if-else logic to the data in a dataframe. How about if-elif-else? Suppose you would like to change any grade 90 and above to ‘A’, any grade between 80-89 to ‘B’, 70-79 to ‘C’, 60-69 to ‘D’, and any grade below 60 to ‘F’.
# change the number grades to letter grades
def convert_grade(x):
if x >= 90:
return 'A'
elif 80<x<89:
return 'B'
elif 70<x<79:
return 'C'
elif 60<x<69:
return 'D'
else:
return 'F'
eng['grade'].apply(convert_grade)
Alice A
Becky B
Cindy F
Dave F
Name: grade, dtype: object
Coding Challenge! < / >
In this coding challenge, we’ll work with the data on the 2022 Marathon.
# get the original data
bm_22
| BibNumber | FullName | SortName | AgeOnRaceDay | Gender | City | StateAbbrev | StateName | Zip | CountryOfResAbbrev | CountryOfResName | CountryOfCtzAbbrev | CountryOfCtzName | OfficialTime | RankOverall | RankOverGender | RankOverDivision | EventGroup | SubGroupLabel | SubGroup | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 4 | Evans Chebet | Chebet, Evans | 33 | M | Kapsabet | NaN | NaN | NaN | KEN | Kenya | KEN | Kenya | 2:06:51 | 1 | 1 | 1 | Runners | NaN | NaN |
| 1 | 5 | Lawrence Cherono | Cherono, Lawrence | 33 | M | Eldoret | NaN | NaN | NaN | KEN | Kenya | KEN | Kenya | 2:07:21 | 2 | 2 | 2 | Runners | NaN | NaN |
| 2 | 1 | Benson Kipruto | Kipruto, Benson | 31 | M | Kapsabet | NaN | NaN | NaN | KEN | Kenya | KEN | Kenya | 2:07:27 | 3 | 3 | 3 | Runners | NaN | NaN |
| 3 | 9 | Gabriel Geay | Geay, Gabriel | 25 | M | Tampa | FL | Florida | 33647 | USA | United States of America | TAN | Tanzania | 2:07:53 | 4 | 4 | 4 | Runners | NaN | NaN |
| 4 | 11 | Eric Kiptanui | Kiptanui, Eric | 31 | M | NaN | NaN | NaN | NaN | KEN | Kenya | KEN | Kenya | 2:08:47 | 5 | 5 | 5 | Runners | NaN | NaN |
| ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... | ... |
| 24829 | 7681 | Paul Kent | Kent, Paul | 58 | M | Hingham | MA | Massachusetts | 2043 | USA | United States of America | USA | United States of America | 6:56:35 | 24830 | 14258 | 1451 | Runners | Para T61-T62 | Para T61-T62 |
| 24830 | 7638 | Aaron Burros | Burros, Aaron | 51 | M | Houston | TX | Texas | 77024 | USA | United States of America | USA | United States of America | 6:58:12 | 24831 | 14259 | 1899 | Runners | NaN | NaN |
| 24831 | 28701 | Julian Martin Arjona | Martin Arjona, Julian | 61 | M | Loja | NaN | NaN | 18300 | ESP | Spain | ESP | Spain | 6:58:50 | 24832 | 14260 | 1056 | Runners | NaN | NaN |
| 24832 | 5398 | Jorge Valenciano | Valenciano, Jorge | 38 | M | Guadalupe | NaN | NaN | 67192 | MEX | Mexico | MEX | Mexico | 7:01:32 | 24833 | 14261 | 4818 | Runners | NaN | NaN |
| 24833 | 2519 | Jason Liddle | Liddle, Jason | 45 | M | Victor | NY | New York | 14564 | USA | United States of America | USA | United States of America | 7:24:39 | 24834 | 14262 | 2298 | Runners | NaN | NaN |
24834 rows × 20 columns
Can you use apply() to update the OfficialTime column such that completion time between 2 and 3 hours are changed to the string ‘Extremely fast’ and completion time between 3 - 4 hours are changed to the string ‘fast’ with all other values unchanged in this column?
Solutions to exercises#
Here are the solutions to some of the exercises in this notebook.
### find out which countries have runners in 2021 but not in 2022
### this solution may look a bit convoluted to you and it's intended to be so
# drop duplicate countries in bm_21 and get two columns after dropping the duplicates
bm_21_ctry = bm_21.drop_duplicates(subset='CountryOfResName')[['CountryOfResName', 'FullName']]
# drop duplicate countries in bm_22 and get the same two columns after dropping the duplicates
bm_22_ctry = bm_22.drop_duplicates(subset='CountryOfResName')[['CountryOfResName', 'FullName']]
# merge the two new dfs on the CountryOfResName column with the merge type being left outer join
bm_merge = bm_21_ctry.merge(bm_22_ctry, how='left', on='CountryOfResName')
# get the rows from the merged df where the non-matching keys in bm_22 get a value of NaN
bm_merge.loc[bm_merge['FullName_y'].isna()==True, 'CountryOfResName']
### find out which countries have runners in 2021 but not in 2022
# this is a non-convoluted solution to the same exercise
bm_21_country = set(bm_21['CountryOfResName'].to_list())
bm_22_country = set(bm_22['CountryOfResName'].to_list())
bm_21_country - bm_22_country
# Use apply() to change the completion time
def convert_time(r):
if r['OfficialTime'].startswith('2'):
r['OfficialTime'] = 'Extremely fast'
elif r['OfficialTime'].startswith('3'):
r['OfficialTime'] = 'Fast'
return r
bm_22.apply(convert_time, axis=1)
# Use lambda function in apply to change the completion time
bm_22['OfficialTime'] = bm_22['OfficialTime'].apply(lambda x: 'Extremely fast' \
if x.startswith('2') \
else ('Fast' if x.startswith('3') else x))
